Why so Sheddy? Tracking how Environmental DNA Production Varies Within a Single Daphnia
Many species are under threat from human activities and climate change in aquatic habitats, including in Southern Ontario, and conservationists need every tool at their disposal to identify where these species can be found and to evaluate the health of at-risk populations.
One new but increasingly important tool for conservationists and researchers alike is environmental DNA or “eDNA.” All organisms regularly release small amounts of DNA into the environment — for example, through excreted waste products, the sloughed scales of a fish or the molted carapace of a crayfish. In an aquatic environment, DNA from these sources eDNA are suspended in the water, allowing keen researchers to identify the presence of a particular species through a simple water sample alone.
There’s no doubt that eDNA has become a valuable tool to detect species of interest. However, conservationists also hope that it can provide the information necessary to estimate how large a population is. Knowing population size is important to assess its overall health. So how well can eDNA predict population size? The first step to answering that question is to figure out how much eDNA just one member of a species produces.
A new study has found that the amount of eDNA shed by an organism can be influenced by multiple factors, and that these factors must be taken into account if using eDNA to estimate population abundance. The research was led by PhD candidate Sharon Wang, Dr. John Fryxell and Dr. Robert Hanner, all in the Department of Integrative Biology.
When evaluating the information gained from eDNA, the researchers knew from the start there would be one major difficulty: eDNA production is related to metabolic rate, and metabolic rate is highly variable throughout the life of an organism.
“There is a lot of variation in metabolic rate among individuals. The magnitude of this variation can in turn have large impacts on eDNA production,” explains Wang. “This means that if we are targeting eDNA, we have to account for how metabolic rate and therefore eDNA production can vary across the life cycle.”
Over its lifetime, an organism grows, reproduces, and dies. Throughout these stages, they may encounter fluctuating temperatures and changing food availability. All of these factors can strongly influence metabolic rate.
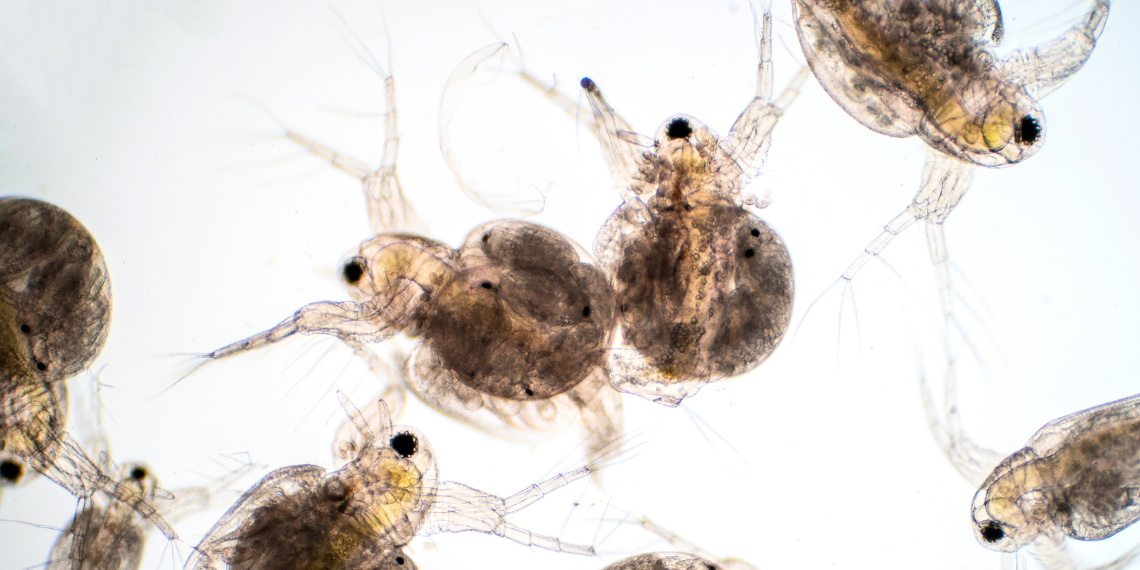
Wang set up an experiment to investigate how life stage, temperature and food availability can impact eDNA production. She used Daphnia magna, a small, planktonic crustacean, that lives naturally in waters throughout Southern Ontario.
Daphnia frequently reproduce by cloning themselves, which allows researchers like Wang to create a whole colony of genetically identical animals.
The Daphnia were split into groups and exposed to different combinations of food availability and temperature which they might encounter in waters around the Great Lakes. Individual Daphnia were held in a vial of freshwater and their eDNA production was measured after 24 hours.
The results showed that Daphnia produced the most eDNA at warmer temperatures, and that larger Daphnia also produced more eDNA. Wang further found that pregnancy and a high level of food availability also increased eDNA production.
“This tells us that it’s a dynamic system and if we want to make any predictive population models using eDNA, we have to consider multiple factors that could contribute to the pattern we found,” says Wang.
For the many species at risk — including important fisheries species — in the waters of Southern Ontario, this means future studies should incorporate indicators of ecology, physiology, and life history if eDNA is to be used as a quantitative conservation tool.
“We did some very interesting work that provides a better understanding of how to interpret eDNA data, but there’s still much more to learn.”
Read the full study in the journal Environmental DNA.
Read about other CBS Research Highlights.